Serotypes of salmonella typhi
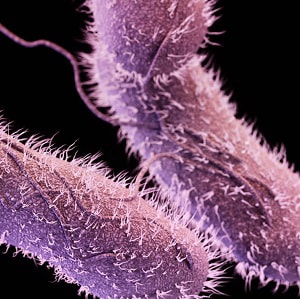
Medical illustration of non-typhoidal Salmonella
Serotypes are groups within a single species of microorganisms, such as bacteria or viruses, which share distinctive surface structures. For instance, Salmonella bacteria look alike under the microscope but can be separated into many serotypes based on two structures on their surface:
- The outermost portion of the bacteria’s surface covering, called the O antigen; and
- A slender threadlike structure, called the H antigen, that is part of the flagella.
The O antigens are distinguished by their different chemical make-up. The H antigens are distinguished by the protein content of the flagella. Each O and H antigen has a unique code number. Scientists determine the serotype based on the distinct combination of O and H antigens.
Salmonella have many different serotypes. Some serotypes are only found in one kind of animal or in a single place. Others are found in many different animals and all over the world. Some can cause especially severe illnesses when they infect people; while others cause milder illnesses.
- The bacteria’s surface are covered with lipopolysaccharide (LPS). The outermost portion of the LPS is the O antigen.
- Flagella are whip-like tails that bacteria use to move around. Flagella is the whole structure, while the slender threadlike portion of the flagella is called the H antigen.

New technology is transforming how we detect and investigate outbreaks. Watch the video to learn more.
Some groups of people, such as older adults, people with weakened immune systems, and children under five years old have a higher risk for Salmonella infection. Infections in these groups can be more severe, resulting in long-term health consequences or death. 1
More than 2,500 serotypes have been described for Salmonella; but, because they are rare, scientists know very little about most of them. Less than 100 serotypes account for most human infections. What we learn about the more common serotypes can help us better understand illness and the natural history of all the Salmonella strains.
Serotyping is a subtyping test based on differences in microbial (e.g., viral or bacterial) surfaces. Serology refers to the antibodies that form because of a viral or bacterial infection. Serotyping is sometimes referred to as serology, but this is technically inaccurate.
Since the 1960s, public health scientists in the US have used serotyping to help find Salmonella outbreaks and track them to their sources. Laboratory experts serotype the Salmonella from infected people. When cases with one serotype increase, they suspect an outbreak and disease detectives start their investigation.
Serotyping has been the core of public health monitoring of Salmonella infections for over 50 years. Now, scientists use DNA testing to further divide each serotype into more subtypes and to detect more outbreaks. With the next generation of sequencing technology, advancements continue as the laboratory can find information about the species, serovar, and subtype of bacteria in just one test. Currently, at least two scientists must generate these three important pieces of information using three separate tests or more. 2
Resistance to two clinically important drugs, ceftriaxone (a cephem) and ciprofloxacin (a quinolone), has climbed in non-typhoidal Salmonella since 1996. In 2011, about 5% of Salmonella tested by CDC were resistant to five or more types of drugs. 3
CDC has posted a series of interactive graphs that allow users to see the percentage of Salmonella human isolates resistant to various antibiotics tracked, by year, through the National Antimicrobial Resistance Monitoring System (NARMS). This graph includes the option to view all Salmonella isolates or any one of five common serotypes with resistance to antibiotics used to treat Salmonella infections: Enteritidis, Typhimurium, Newport, Heidelberg, and I 4,[5],12:i: -.
Salmonellosis is a major cause of bacterial enteric illness in both humans and animals. Each year an estimated 1.4 million cases of salmonellosis occur among humans in the United States (15). In approximately 35,000 of these cases, Salmonella isolates are serotyped by public health laboratories and the results are electronically transmitted to the Centers for Disease Control and Prevention (CDC). This information is used by local and state health departments and CDC to monitor local, regional, and national trends in human salmonellosis and to identify possible outbreaks of salmonellosis (1, 5). Over the past 25 years, the National Salmonella Surveillance System has provided valuable information on the incidence of human salmonellosis in the United States and trends in specific serotypes. The recently implemented Salmonella Outbreak Detection Algorithm, another valuable tool for the recognition of outbreaks (11), allows users to detect increases in human infections due to specific Salmonella serotypes. Salmonella surveillance activities depend upon the accuracy of serotype identification and are facilitated by standardized nomenclature. The National Salmonella Reference Laboratory at CDC assists public health laboratories in the United States in serotype identification by providing procedure manuals, training workshops, updates, and assistance with the identification of problem isolates.
There are currently 2,463 serotypes (serovars) of Salmonella (18). The antigenic formulae of Salmonella serotypes are defined and maintained by the World Health Organization (WHO) Collaborating Centre for Reference and Research on Salmonella at the Pasteur Institute, Paris, France (WHO Collaborating Centre), and new serotypes are listed in annual updates of the Kauffmann-White scheme (18, 19).
Salmonella nomenclature is complex, and scientists use different systems to refer to and communicate about this genus. However, uniformity in Salmonella nomenclature is necessary for communication between scientists, health officials, and the public. Unfortunately, current usage often combines several nomenclatural systems that inconsistently divide the genus into species, subspecies, subgenera, groups, subgroups, and serotypes (serovars), and this causes confusion. CDC receives many inquiries concerning the appropriate Salmonella nomenclature for the reporting of results and for use in scientific publications.
The nomenclature for the genus Salmonella has evolved from the initial one serotype-one species concept proposed by Kauffmann (12) on the basis of the serologic identification of O (somatic) and H (flagellar) antigens. Each serotype was considered a separate species (for example, S. paratyphi A, S. newport, and S. enteritidis); this concept, if used today, would result in 2,463 species of Salmonella. Other taxonomic proposals have been based on the clinical role of a strain, on the biochemical characteristics that divide the serotypes into subgenera, and ultimately, on genomic relatedness. The proposals for nomenclature changes in the genus have been summarized previously (8, 9, 14).
The defining development in Salmonella taxonomy occurred in 1973 when Crosa et al. (6) demonstrated by DNA-DNA hybridization that all serotypes and subgenera I, II, and IV of Salmonella and all serotypes of “Arizona” were related at the species level; thus, they belonged in a single species. The single exception, subsequently described, is S. bongori, previously known as subspecies V, which by DNA-DNA hybridization is a distinct species (21). Since S. choleraesuis appeared on the Approved List of Bacterial Names (23) as the type species of Salmonella, it had priority as the species name. The name “choleraesuis,” however, refers to both a species and a serotype, which causes confusion. In addition, the serotype Choleraesuis is not representative of the majority of serotypes because it is biochemically distinct, being arabinose and trehalose negative (4, 13).
In 1986 the Subcommittee of Enterobacteriaceae of the International Committee on Systematic Bacteriology at the XIV International Congress of Microbiology unanimously recommended that the type species for Salmonella be changed to S. enterica (17), a name coined by Kauffmann and Edwards in 1952 (13), because no serotype shares this name. In 1987, Le Minor and Popoff of the WHO Collaborating Centre formally made a proposal as a “Request for an Opinion” to the Judicial Commission of the International Committee of Systematic Bacteriology (14). The recommendation was adopted by CDC, by Ewing in 1986 in the 4th edition of Edward's and Ewing's Identification of Enterobactericeae (8), and by other laboratories (16).
Nonetheless, the request was denied by the Judicial Commission. Although the Judicial Commission was generally in favor of S. enterica as the type species of Salmonella, its members believed that the status of Salmonella serotype Typhi, the causative agent of typhoid fever, was not adequately addressed in this request for an opinion. They were concerned that if S. enterica were adopted as the type species, Salmonella serotype Typhi would be referred to as S. enterica subsp. enterica serotype Typhi and might be missed or overlooked by physicians in the same way that S. choleraesuis subsp. choleraesuis serotype Typhi might be overlooked. From this perspective, nothing would be gained by changing the type species name. The Judicial Commission therefore ruled that S. choleraesuis be retained as the legitimate type species pending an amended request for an opinion (24). To comply with this ruling, in 1999 Euzéby (7) made an amended request, which is pending, to adopt S. enterica as the type species of Salmonella while retaining the species “S. typhi” as an exception.
In 1987, Le Minor and Popoff (14) also proposed that the seven subgenera of Salmonella be referred to as subspecies (subspecies I, II, IIIa, IIIb, IV, V, and VI). Subgenus III was divided into IIIa and IIIb by genomic relatedness and biochemical reactions. Subspecies IIIa (S. enterica subsp. arizonae) includes the monophasic “Arizona” serotypes and subspecies IIIb (S. enterica subsp. diarizonae) contains the diphasic serotypes. All “Arizona” serotypes had been incorporated into the Kauffmann-White scheme by Rohde in 1979 (22).
THE CURRENT SYSTEM USED BY CDC
In this report we update the nomenclature used at CDC for members of the genus Salmonella. The nomenclatural system is based on recommendations from the WHO Collaborating Centre and is summarized in Tables Tables1, 1 , ,2, 2 , and and3 3 .
Salmonella species, subspecies, serotypes, and their usual habitats, Kauffmann-White scheme a
Current system of Salmonella nomenclature
used by WHO, CDC, and ASM
Salmonella is named after an American bacteriologist, D. E. Salmon, who first isolated Salmonella choleraesuis from porcine intestine in 1884 (1). Salmonellae are a group of bacterial organisms with a high genetic similarity and are differentiated by their serotyping results. The antigenic classification system of various Salmonella serotypes used today is a result accumulated from many years of studies on antibody interactions with surface antigens of Salmonella organisms established by Kauffman and White almost a century ago. All antigenic formulae of recognized Salmonella serotypes are listed in a document called the Kauffmann-White scheme (2). The World Health Organization Collaborating Centre for Reference and Research on Salmonella at the Pasteur Institute, Paris, France (WHO Collaborating Centre) is responsible for the updating of the scheme. Every year newly recognized serotypes are reported in the Research in Microbiology by Popoff et al. and in the latest report published in 2004, there are a total of 2,541 serotypes in the genus Salmonella (3). The terms “serotype” and “serovar” are both frequently used, but according to the Rules of the Bacteriological Code (1990 Revision) established by the Judicial Commission of the International Committee on the Systematics of Prokaryotes, the term serovar is preferred to the term serotype. Thus “serovar” is used in the Kauffmann-White scheme.
Salmonella nomenclature is complex and still evolving. Currently, the nomenclature system used at the Centers for Disease Control and Prevention (CDC) for the genus Salmonella is based on recommendations from the WHO Collaborating Centre. According to the CDC system, the genus Salmonella contains two species, S. enterica, the type species, and S. bongori. On March 18, 2005, a new species, “Salmonella subterranean” was validly approved by the Judicial Commission (4); CDC may incorporate the species in their system in the near future. S. enterica consists of six subspecies (2,5): I, S. enterica subsp. enterica; II, S. enterica subsp. salamae; IIIa, S. enterica subsp. arizonae; IIIb, S. enterica subsp. diarizonae; IV, S. enterica subsp. houtenae; and VI, S. enterica subsp. indica.
In subspecies I, serotypes (or serovars) are designated by a name usually indicative of the associated diseases, their geographic origins, or their usual habitats. In the remaining subspecies as well as those of S. bongore, antigenic formulae determined according to the Kauffmann-White scheme (2) are used for those unnamed serotypes. Some members of these subspecies may have been named before 1966 and thus their names are retained and cited as those in the subspecies I.
To avoid confusion between serotypes and species, the serotype name is not italicized and starts with a capital letter. When cited at the first time in a report, the genus name is given followed by the word “serotype” (or the abbreviation “ser.”) and then the serotype name, e.g., Salmonella serotype or ser. Choleraesuis, and Salmonella serotype or ser. Typhi. Afterward the name may be shortened with the genus name followed directly by the serotype name, e.g., Salmonella Choleraesuis or S. Choleraesuis, and Salmonella Typhi or S. Typhi (3). Because the type species name, enterica, has not been approved before 2005, serotype names are used directly after the genus name without the mention of the species. Following the officially approval of “enterica” as the type species name from January 2005 (6), further amendment to include species name in the Salmonella nomenclature by CDC may be expected.
For those designated by their antigenic formulae, the subspecies name is written in Roman letters (not italicized) followed by their antigenic formulae, including O (somatic) antigens, H (flagellar) antigens (phase 1), and H antigens (phase 2, if present). A colon is used in between each antigen, e.g., Salmonella serotype II 39:z 10 : z 6 . For serotypes in S. bongori (previously belongs to subgenous V), V is still used for consistency, e.g., S. V 13,22:z 35 :- (3).
In publications of the American Society for Microbiology (ASM), the Salmonella nomenclature used at CDC was accepted as the standard form by the Publications Board in the 1999 meeting and the Instruction to Authors has been updated to include this information since 2000 (7). The 2006 ASM Instruction to Authors indicated that, for the species, “Salmonella enterica” is used at the first time, and “S. enterica” thereafter; for the subspecies, “Salmonella enterica subsp. arizonae” is used at first, and “S. enterica subsp. arizonae” thereafter. Serotype names should be in Roman type with the first letter capitalized, e.g., Salmonella enterica serotype Typhimurium. After the first use, the serotype may be used without a species name, e.g., Salmonella serotype Typhimurium.
1. Smith T. The hog-cholera group of bacteria. U.S. Bur Anim Ind Bull 1894; 6:6-40.
2. Popoff MY, Le Minor L. Antigenic formulas of the Salmonella serovars, 8th revision, World Health Organization Collaborating Centre for Reference and Research on Salmonella, Pasteur Institute, Paris, France. 2001.
3. Popoff MY, Bockemьhl J, Gheesling LL. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res Microbiol 2004; 155:568-570.
4. Shelobolina ES, Sullivan SA, O'neill KR, Nevin KP, Lovley DR. Isolation, characterization, and U(VI)-reducing potential of a facultatively anaerobic, acid-resistant bacterium from low-pH, nitrate- and U(VI)-contaminated subsurface sediment and description of Salmonella subterranea sp. nov. Appl Environ Microbiol 2004; 70:2959-2965.
5. Brenner FW, McWhorter-Murlin AC. Identification and serotyping of Salmonella. Centers for Disease Control and Prevention, Atlanta, Ga. 1998.
6. Judicial Commission of the International Committee on Systematics of Prokaryotes. The type species of the genus Salmonella Lignieres 1900 is Salmonella enterica (ex Kauffmann and Edwards 1952) Le Minor and Popoff 1987, with the type strain LT2T, and conservation of the epithet enterica in Salmonella enterica over all earlier epithets that may be applied to this species. Opinion 80. Int J Syst Evol Microbiol 2005; 55:519-520.
7. Publications Board. Publications Board meeting minutes. Salmonella nomenclature. ASM News 1999; 65:769.
Related terms:
Download as PDF
About this page
Malaria (Plasmodium Species)
Salmonella typhi and Salmonella paratyphi can be acquired in developing countries worldwide. As for malaria, patients with enteric fever may present with fever, headache, nausea, malaise, anorexia, and myalgias. Prominent gastrointestinal symptoms (abdominal pain, constipation, or diarrhea), the findings of rose spots or relative bradycardia, and a history of unsanitary food or water consumption may help to support a diagnosis of enteric fever. A history of prior vaccination against S. typhi may not be useful in ruling out enteric fever because it is only 50% to 80% effective and does not protect against paratyphoidal illness.
Typhoid and Paratyphoid (Enteric) Fever
Salmonella Typhi, Salmonella Paratyphi A, and Salmonella Paratyphi B are serovars within the genus Salmonella of the family Enterobacteriaceae. These Gram-negative, motile bacilli do not ferment lactose.
Only two species are currently recognized within the genus Salmonella, Salmonella enterica and S. bongori, and only the former is important with respect to human disease. There are six subspecies of S. enterica, of which subspecies I, S. enterica subspecies enterica, contains all the important pathogens that cause human disease. S. enterica subspecies enterica is further subdivided into >2500 serovars (i.e., distinct serotypes) based on specific somatic O antigens, capsular polysaccharide Vi antigen, and flagellar H antigens expressed by the organism. The antigenic serotyping scheme (Kauffman–White scheme) defines a serovar by its O polysaccharide antigens (and also whether capsular polysaccharide Vi is expressed) and its H flagellar antigens. The serogroup of a Salmonella is defined by its O antigens, while serovar is defined by the full antigenic structure that includes the flagellar antigens and whether Vi is expressed. O antigens are part of the lipopolysaccharide (LPS) of the bacterial outer membrane. The lipid A (endotoxin) portion of LPS is a glucosamine-based phospholipid that makes up the outer monolayer of the bacterial outer membrane. Attached to lipid A is a core polysaccharide that is essentially identical in all the important Salmonella serovars that cause human disease, particularly invasive disease. The most external surface component attached to the core polysaccharide is an O polysaccharide that consists of terminal O repeat units linked one to another. This is exposed to the environment in Salmonella Paratyphi A and B, whereas in Salmonella Typhi the capsular Vi (for “virulence”) polysaccharide (homopolymer of N-acetylgalacturonic acid) 7 covers the O polysaccharide.
The terminal O polysaccharide of Salmonella varies in structure depending on the sugars comprising the core unit and their linkages one to another. Salmonella Paratyphi A falls into serogroup A, Salmonella Paratyphi B into serogroup B, and Salmonella Typhi into serogroup D. The O repeat units of Salmonella serogroups A, B and D share a common trisaccharide backbone that consists of repeats of mannose, rhamnose, and galactose. 8 Attached to the backbone is another dideoxyhexose sugar that is α-1,3-linked to the mannose residue. 8 If the α-1,3-linked dideoxyhexose sugar is abequose, the resultant structure constitutes immunodominant antigen “4” that defines O serogroup B; if the sugar is paratose, the structure creates immunodominant antigen “2” that defines serogroup A; if the sugar is tyvelose, immunodominant antigen “9” is created that defines O serogroup D.
Some Salmonella express two different antigenic forms of flagella, called phase 1 and phase 2. Salmonella Typhi and Paratyphi A express only phase 1 flagellar antigens, H:a and H:d, respectively, while Salmonella Paratyphi B expresses both phase 1 flagella H:b and phase 2 flagella H:1,2. Bacteriologic confirmation of Salmonella Typhi, Salmonella Paratyphi A, and Salmonella Paratyphi B can be made by agglutination with typing sera or by multiplex polymerase chain reaction (PCR). 9
Whereas other pathogenic Enterobacteriaceae such as Shigella and non-typhoidal Salmonella stably carry R factor plasmids encoding antibiotic resistance, until about 1990 this was the exception with typhoid bacilli. The first antibiotic used to treat typhoid fever, chloramphenicol, reported in 1948, 10 was immensely useful for a quarter century thereafter (and remains useful where strains of Salmonella Typhi remain susceptible). However, rather suddenly, large-scale epidemics of chloramphenicol-resistant typhoid fever ensued, first in Mexico (1972) 11,12 and then in Southeast Asia (1974). 13 After ∼2 years in Mexico, the resistant strain disappeared and was replaced by chloramphenicol-sensitive strains. In 1979 and 1980, chloramphenicol-resistant typhoid appeared in Lima, Peru 14 but these resistant strains also disappeared after a few years and were replaced by chloramphenicol-sensitive Salmonella Typhi. In these instances the antibiotic-resistance genes were encoded on plasmids of incompatibility group HI1. 11,14 Beginning in the late 1980s, Salmonella Typhi strains resistant to chloramphenicol, amoxicillin, and trimethoprim-sulfamethoxazole disseminated widely throughout Asia. 15–17 Alternative effective antibiotics included oral ciprofloxacin and parenteral ceftriaxone; however, the widespread use of ciprofloxacin and other fluoroquinolones, often in inadequate dosages and duration, encouraged the emergence of fluoroquinolone-resistant strains. 18
The full sequences of the genome of two isolates of Salmonella Typhi have been reported including modern multiresistant strain CT18 from Vietnam and venerable strain Ty2, isolated in Russia in about 1915. The genome of the former has 4 809 037 base pairs (bp) and 4599 open reading frames (ORFs), 19 while Ty2 has 4 791 961 bp and 4399 ORFs. 20 There is 98% genomic homology between CT18 and Ty2. A striking revelation was that these genomes have undergone degradation (compared to Salmonella Typhimurium); each genome shows >200 pseudogenes, 195 of which are identical. There are also segments of genome present in Typhimurium that are not evident in Typhi. Since more than a dozen of the pseudogenes encode fimbrial attachment factors, loss of these likely explains the narrow human host specificity of Typhi compared to Typhimurium.
Salmonella Typhi also exhibits 10 Salmonella pathogenicity islands including ones involved in cell invasion (SPI1), intracellular survival (SPI2) and Vi biosynthesis (SPI7). There is >80% sequence homology with Salmonella Typhimurium.
Salmonella Paratyphi A has a 4 585 229 bp genome with 4263 ORFs and considerable homology with Salmonella Typhi. 21 Paratyphi A also manifests genomic degradation, with 173 pseudogenes (28 identical to Typhi). 21 Salmonella Paratyphi A followed a distinct evolutionary path to evolve into a pathogen exhibiting a similar pathogenesis and host specificity as Typhi.
Salmonella Typhi and Salmonella Paratyphi A
Kenneth E. Sanderson, . Randal N. Johnston, in Molecular Medical Microbiology (Second Edition) , 2015
Salmonella Typhi and Salmonella Paratyphi A, collectively known as typhoidal Salmonella, are causal agents for a serious, invasive (bacteraemic), sometimes fatal disease of humans called typhoid fever or paratyphoid fever (also called enteric fevers). Salmonella Typhi, the lineage causing typhoid fever, is the main group; while Salmonella Paratyphi A, the lineage causing paratyphoid fever, belongs to the second group which comprises a set of three paratyphoid types (the other two being S. Paratyphi C and d-tartrate-negative S. Paratyphi B). All of these lineages are adapted to humans, with S. Typhi and S. Paratyphi A being strictly restricted to growth in humans, and S. Paratyphi C being able to establish infections in experimental animals quite easily (at moderate infection doses); the host-restriction status of d-tartrate-negative S. Paratyphi B is so far unclear. Representing an update on the version published in the first edition of this book that very thoroughly summarized the knowledge available at that time, most of the emphasis in this chapter is on typhoid fever due to S. Typhi in relation to its taxonomy, genomics and genetics, diagnosis, association with disease, mechanisms of invasion and pathogenesis, and antibiotic and vaccine strategies to minimize its impact. The impact of typhoidal Salmonella on human hosts is indeed very large. In 2000, typhoid fever caused over 20 million illnesses and more than 200 000 deaths, whereas paratyphoid fever caused an estimated 5.4 million illnesses worldwide. The greatest burden of illness was suffered by infants, children and adolescents in south-central and south-eastern Asia. Typhoid and paratyphoid fever usually present as clinically similar acute febrile illnesses; accurate diagnosis relies on confirmation by laboratory tests. Paratyphoid fever is usually the result of infection by S. Paratyphi A, and recent reports show an increasing incidence of S. Paratyphi A causing enteric fever in developing counties in Asia and show that the earlier notion that paratyphoid fever is less serious than typhoid fever is not correct. The organism must be cultured and identified to make a clear diagnosis; clinical symptoms alone are not adequate. Culture from blood is less sensitive than bone marrow culture and often gives negative results even when bone marrow cultures are positive, but it is usually the practical first choice for patient diagnosis and for epidemiologic studies of the burden of typhoid and paratyphoid fever. Because of a lack of good diagnostic tools, and because the sites of endemic disease are often deficient in clinical and laboratory facilities, the extent of the burden of enteric fever is often poorly characterized in much of the world, especially in sub-Saharan Africa.
Key facts
- Salmonella is 1 of 4 key global causes of diarrhoeal diseases.
- Most cases of salmonellosis are mild; however, sometimes it can be life-threatening. The severity of the disease depends on host factors and the serotype of Salmonella.
- Antimicrobial resistance is a global public health concern and Salmonella is one of the microorganisms in which some resistant serotypes have emerged, affecting the food chain.
- Basic food hygiene practices, such as "cook thoroughly", are recommended as a preventive measure against salmonellosis.
Salmonella is a gram negative rods genus belonging to the Enterobacteriaceae family. Within 2 species, Salmonella bongori and Samonella enterica, over 2500 different serotypes or serovars have been identified to date. Salmonella is a ubiquitous and hardy bacteria that can survive several weeks in a dry environment and several months in water.
While all serotypes can cause disease in humans, a few are host-specific and can reside in only one or a few animal species: for example, Salmonella enterica serotype Dublin in cattle and Salmonella enterica serotype Choleraesuis in pigs. When these particular serotypes cause disease in humans, it is often invasive and can be life-threatening. Most serotypes, however, are present in a wide range of hosts. Typically, such serotypes cause gastroenteritis, which is often uncomplicated and does not need treatment, but disease can be severe in the young, the elderly, and patients with weakened immunity. This group features Salmonella enterica serotype Enteritidis and Salmonella enterica serotype Typhimurium, the two most important serotypes of Salmonella transmitted from animals to humans in most parts of the world.
The disease
Salmonellosis is a disease caused by the bacteria Salmonella. It is usually characterized by acute onset of fever, abdominal pain, diarrhoea, nausea and sometimes vomiting.
The onset of disease symptoms occurs 6–72 hours (usually 12–36 hours) after ingestion of Salmonella, and illness lasts 2–7 days.
Symptoms of salmonellosis are relatively mild and patients will make a recovery without specific treatment in most cases. However, in some cases, particularly in children and elderly patients, the associated dehydration can become severe and life-threatening.
Although large Salmonella outbreaks usually attract media attention, 60–80% of all salmonellosis cases are not recognized as part of a known outbreak and are classified as sporadic cases, or are not diagnosed as such at all.
Sources and transmission
- Salmonella bacteria are widely distributed in domestic and wild animals. They are prevalent in food animals such as poultry, pigs, and cattle; and in pets, including cats, dogs, birds, and reptiles such as turtles.
- Salmonella can pass through the entire food chain from animal feed, primary production, and all the way to households or food-service establishments and institutions.
- Salmonellosis in humans is generally contracted through the consumption of contaminated food of animal origin (mainly eggs, meat, poultry, and milk), although other foods, including green vegetables contaminated by manure, have been implicated in its transmission.
- Person-to-person transmission can also occur through the faecal-oral route.
- Human cases also occur where individuals have contact with infected animals, including pets. These infected animals often do not show signs of disease.
Treatment
Treatment in severe cases is electrolyte replacement (to provide electrolytes, such as sodium, potassium and chloride ions, lost through vomiting and diarrhoea) and rehydration.
Routine antimicrobial therapy is not recommended for mild or moderate cases in healthy individuals. This is because antimicrobials may not completely eliminate the bacteria and may select for resistant strains, which subsequently can lead to the drug becoming ineffective. However, health risk groups such as infants, the elderly, and immunocompromised patients may need to receive antimicrobial therapy. Antimicrobials are also administered if the infection spreads from the intestine to other body parts. Because of the global increase of antimicrobial resistance, treatment guidelines should be reviewed on a regular basis taking into account the resistance pattern of the bacteria based on the local surveillance system.
Prevention methods
Prevention requires control measures at all stages of the food chain, from agricultural production, to processing, manufacturing and preparation of foods in both commercial establishments and at home.
Preventive measures for Salmonella in the home are similar to those used against other foodborne bacterial diseases (see recommendations for food handlers below).
The contact between infants/young children and pet animals that may be carrying Salmonella (such as cats, dogs, and turtles) needs careful supervision.
National and regional surveillance systems on foodborne diseases are important means to know and follow the situation of these diseases and also to detect and respond to salmonellosis and other enteric infections in early stages, and thus to prevent them from further spreading.
Recommendations for the public and travellers
The following recommendations will help ensure safety while travelling:
- Ensure food is properly cooked and still hot when served.
- Avoid raw milk and products made from raw milk. Drink only pasteurized or boiled milk.
- Avoid ice unless it is made from safe water.
- When the safety of drinking water is questionable, boil it or if this is not possible, disinfect it with a reliable, slow-release disinfectant agent (usually available at pharmacies).
- Wash hands thoroughly and frequently using soap, in particular after contact with pets or farm animals, or after having been to the toilet.
- Wash fruits and vegetables carefully, particularly if they are eaten raw. If possible, vegetables and fruits should be peeled.
- A guide on safe food for travellers
Recommendations for food handlers
WHO provides the following guidance for people handling food:
- Both professional and domestic food handlers should be vigilant while preparing food and should observe hygienic rules of food preparation.
- Professional food handlers who suffer from fever, diarrhoea, vomiting or visible infected skin lesions should report to their employer immediately.
- The WHO Five keys to safer food serve as the basis for educational programmes to train food handlers and educate consumers. They are especially important in preventing food poisoning. The five keys to Safer Food are:
- keep clean
- separate raw and cooked
- cook thoroughly
- keep food at safe temperatures
- use safe water and raw materials.
- Five keys to safer food
Recommendations for producers of fruits, vegetables and fish
The WHO Five keys to growing safer fruits and vegetables: promoting health by decreasing microbial contamination and the Five keys to safer aquaculture products to protect public health provide rural workers, including small farmers who grow fresh fruits and vegetables and fish for themselves, their families and for sale in local market with key practices to prevent microbial contamination.
The Five keys to growing safer fruits and vegetables are:
- Practice good personal hygiene.
- Protect fields from animal faecal contamination.
- Use treated faecal waste.
- Evaluate and manage risks from irrigation water.
- Keep harvest and storage equipment clean and dry.
- Five keys to growing safer fruits and vegetables
The Five keys to safer aquaculture products to protect public health are:
- Practice good personal hygiene.
- Clean the pond site.
- Manage water quality.
- Keep fish healthy.
- Use clean harvest equipment and containers.
- Five keys to safer aquaculture products to protect public health
WHO response
In partnership with other stakeholders, WHO is strongly advocating the importance of food safety as an essential element in ensuring access to safe and nutritious diets. WHO is providing policies and recommendations that cover the entire food chain from production to consumption, making use of different types of expertise across different sectors.
WHO is working towards the strengthening of food safety systems in an increasingly globalized world. Setting international food safety standards, enhancing disease surveillance, educating consumers and training food handlers in safe food handling are amongst the most critical interventions in the prevention of foodborne illnesses.
WHO is strengthening the capacities of national and regional laboratories in the surveillance of foodborne pathogens, such as Campylobacter and Salmonella.
WHO is also promoting the integrated surveillance of antimicrobial resistance of pathogens in the food chain, collecting samples from humans, food and animals and analysing data across the sectors.
WHO, jointly with FAO, is assisting Member States by coordinating international efforts for early detection and response to foodborne disease outbreaks through the network of national authorities in Member States.
WHO also provides scientific assessments as basis for international food standards, guidelines and recommendations developed by the FAO/WHO Codex Alimentarius Commission to prevent foodborne diseases.
Читайте также:


