Salmonella enterica serovar paratyphi
Related terms:
Download as PDF
About this page
Malaria (Plasmodium Species)
Salmonella typhi and Salmonella paratyphi can be acquired in developing countries worldwide. As for malaria, patients with enteric fever may present with fever, headache, nausea, malaise, anorexia, and myalgias. Prominent gastrointestinal symptoms (abdominal pain, constipation, or diarrhea), the findings of rose spots or relative bradycardia, and a history of unsanitary food or water consumption may help to support a diagnosis of enteric fever. A history of prior vaccination against S. typhi may not be useful in ruling out enteric fever because it is only 50% to 80% effective and does not protect against paratyphoidal illness.
Typhoid and Paratyphoid (Enteric) Fever
Salmonella Typhi, Salmonella Paratyphi A, and Salmonella Paratyphi B are serovars within the genus Salmonella of the family Enterobacteriaceae. These Gram-negative, motile bacilli do not ferment lactose.
Only two species are currently recognized within the genus Salmonella, Salmonella enterica and S. bongori, and only the former is important with respect to human disease. There are six subspecies of S. enterica, of which subspecies I, S. enterica subspecies enterica, contains all the important pathogens that cause human disease. S. enterica subspecies enterica is further subdivided into >2500 serovars (i.e., distinct serotypes) based on specific somatic O antigens, capsular polysaccharide Vi antigen, and flagellar H antigens expressed by the organism. The antigenic serotyping scheme (Kauffman–White scheme) defines a serovar by its O polysaccharide antigens (and also whether capsular polysaccharide Vi is expressed) and its H flagellar antigens. The serogroup of a Salmonella is defined by its O antigens, while serovar is defined by the full antigenic structure that includes the flagellar antigens and whether Vi is expressed. O antigens are part of the lipopolysaccharide (LPS) of the bacterial outer membrane. The lipid A (endotoxin) portion of LPS is a glucosamine-based phospholipid that makes up the outer monolayer of the bacterial outer membrane. Attached to lipid A is a core polysaccharide that is essentially identical in all the important Salmonella serovars that cause human disease, particularly invasive disease. The most external surface component attached to the core polysaccharide is an O polysaccharide that consists of terminal O repeat units linked one to another. This is exposed to the environment in Salmonella Paratyphi A and B, whereas in Salmonella Typhi the capsular Vi (for “virulence”) polysaccharide (homopolymer of N-acetylgalacturonic acid) 7 covers the O polysaccharide.
The terminal O polysaccharide of Salmonella varies in structure depending on the sugars comprising the core unit and their linkages one to another. Salmonella Paratyphi A falls into serogroup A, Salmonella Paratyphi B into serogroup B, and Salmonella Typhi into serogroup D. The O repeat units of Salmonella serogroups A, B and D share a common trisaccharide backbone that consists of repeats of mannose, rhamnose, and galactose. 8 Attached to the backbone is another dideoxyhexose sugar that is α-1,3-linked to the mannose residue. 8 If the α-1,3-linked dideoxyhexose sugar is abequose, the resultant structure constitutes immunodominant antigen “4” that defines O serogroup B; if the sugar is paratose, the structure creates immunodominant antigen “2” that defines serogroup A; if the sugar is tyvelose, immunodominant antigen “9” is created that defines O serogroup D.
Some Salmonella express two different antigenic forms of flagella, called phase 1 and phase 2. Salmonella Typhi and Paratyphi A express only phase 1 flagellar antigens, H:a and H:d, respectively, while Salmonella Paratyphi B expresses both phase 1 flagella H:b and phase 2 flagella H:1,2. Bacteriologic confirmation of Salmonella Typhi, Salmonella Paratyphi A, and Salmonella Paratyphi B can be made by agglutination with typing sera or by multiplex polymerase chain reaction (PCR). 9
Whereas other pathogenic Enterobacteriaceae such as Shigella and non-typhoidal Salmonella stably carry R factor plasmids encoding antibiotic resistance, until about 1990 this was the exception with typhoid bacilli. The first antibiotic used to treat typhoid fever, chloramphenicol, reported in 1948, 10 was immensely useful for a quarter century thereafter (and remains useful where strains of Salmonella Typhi remain susceptible). However, rather suddenly, large-scale epidemics of chloramphenicol-resistant typhoid fever ensued, first in Mexico (1972) 11,12 and then in Southeast Asia (1974). 13 After ∼2 years in Mexico, the resistant strain disappeared and was replaced by chloramphenicol-sensitive strains. In 1979 and 1980, chloramphenicol-resistant typhoid appeared in Lima, Peru 14 but these resistant strains also disappeared after a few years and were replaced by chloramphenicol-sensitive Salmonella Typhi. In these instances the antibiotic-resistance genes were encoded on plasmids of incompatibility group HI1. 11,14 Beginning in the late 1980s, Salmonella Typhi strains resistant to chloramphenicol, amoxicillin, and trimethoprim-sulfamethoxazole disseminated widely throughout Asia. 15–17 Alternative effective antibiotics included oral ciprofloxacin and parenteral ceftriaxone; however, the widespread use of ciprofloxacin and other fluoroquinolones, often in inadequate dosages and duration, encouraged the emergence of fluoroquinolone-resistant strains. 18
The full sequences of the genome of two isolates of Salmonella Typhi have been reported including modern multiresistant strain CT18 from Vietnam and venerable strain Ty2, isolated in Russia in about 1915. The genome of the former has 4 809 037 base pairs (bp) and 4599 open reading frames (ORFs), 19 while Ty2 has 4 791 961 bp and 4399 ORFs. 20 There is 98% genomic homology between CT18 and Ty2. A striking revelation was that these genomes have undergone degradation (compared to Salmonella Typhimurium); each genome shows >200 pseudogenes, 195 of which are identical. There are also segments of genome present in Typhimurium that are not evident in Typhi. Since more than a dozen of the pseudogenes encode fimbrial attachment factors, loss of these likely explains the narrow human host specificity of Typhi compared to Typhimurium.
Salmonella Typhi also exhibits 10 Salmonella pathogenicity islands including ones involved in cell invasion (SPI1), intracellular survival (SPI2) and Vi biosynthesis (SPI7). There is >80% sequence homology with Salmonella Typhimurium.
Salmonella Paratyphi A has a 4 585 229 bp genome with 4263 ORFs and considerable homology with Salmonella Typhi. 21 Paratyphi A also manifests genomic degradation, with 173 pseudogenes (28 identical to Typhi). 21 Salmonella Paratyphi A followed a distinct evolutionary path to evolve into a pathogen exhibiting a similar pathogenesis and host specificity as Typhi.
Salmonella Typhi and Salmonella Paratyphi A
Kenneth E. Sanderson, . Randal N. Johnston, in Molecular Medical Microbiology (Second Edition) , 2015
Salmonella Typhi and Salmonella Paratyphi A, collectively known as typhoidal Salmonella, are causal agents for a serious, invasive (bacteraemic), sometimes fatal disease of humans called typhoid fever or paratyphoid fever (also called enteric fevers). Salmonella Typhi, the lineage causing typhoid fever, is the main group; while Salmonella Paratyphi A, the lineage causing paratyphoid fever, belongs to the second group which comprises a set of three paratyphoid types (the other two being S. Paratyphi C and d-tartrate-negative S. Paratyphi B). All of these lineages are adapted to humans, with S. Typhi and S. Paratyphi A being strictly restricted to growth in humans, and S. Paratyphi C being able to establish infections in experimental animals quite easily (at moderate infection doses); the host-restriction status of d-tartrate-negative S. Paratyphi B is so far unclear. Representing an update on the version published in the first edition of this book that very thoroughly summarized the knowledge available at that time, most of the emphasis in this chapter is on typhoid fever due to S. Typhi in relation to its taxonomy, genomics and genetics, diagnosis, association with disease, mechanisms of invasion and pathogenesis, and antibiotic and vaccine strategies to minimize its impact. The impact of typhoidal Salmonella on human hosts is indeed very large. In 2000, typhoid fever caused over 20 million illnesses and more than 200 000 deaths, whereas paratyphoid fever caused an estimated 5.4 million illnesses worldwide. The greatest burden of illness was suffered by infants, children and adolescents in south-central and south-eastern Asia. Typhoid and paratyphoid fever usually present as clinically similar acute febrile illnesses; accurate diagnosis relies on confirmation by laboratory tests. Paratyphoid fever is usually the result of infection by S. Paratyphi A, and recent reports show an increasing incidence of S. Paratyphi A causing enteric fever in developing counties in Asia and show that the earlier notion that paratyphoid fever is less serious than typhoid fever is not correct. The organism must be cultured and identified to make a clear diagnosis; clinical symptoms alone are not adequate. Culture from blood is less sensitive than bone marrow culture and often gives negative results even when bone marrow cultures are positive, but it is usually the practical first choice for patient diagnosis and for epidemiologic studies of the burden of typhoid and paratyphoid fever. Because of a lack of good diagnostic tools, and because the sites of endemic disease are often deficient in clinical and laboratory facilities, the extent of the burden of enteric fever is often poorly characterized in much of the world, especially in sub-Saharan Africa.
| This is a curated page. Report corrections to Microbewiki. |
Contents
Etiology/Bacteriology
Salmonella enterica are motile, non-lactose fermenting, non-spore forming, gram-negative, rod-shaped bacterium. Salmonella enterica have the ability to ferment glucose resulting in the production of acid and gas [1] . Within the subspecies, enterica, there are three serotypes; Paratyphi A, B, and C. These serotypes are human pathogens that cause paratyphoid fever. Paratyphis A and B are responsible for more cases of disease than infection from Paratyphi C. In the United States, paratyphoid fever is relatively uncommon, while, an estimated 5.4 million outbreaks occurred in East Asia in 2000. Salmonella enterica serovar Paratyphi, also referred to as Salmonella Paratyphi, causes 3% of invasive Salmonella infections in the U.S and is correlated to poor sanitation and lack of clean drinking water. Prevention entails basic sanitation and education [2]. Symptoms of paratyphoid fever typically include fever peaking around 40°C/, red rash covering the trunk, constipation, severe stomach pain, and loss of appetite. Diagnosing paratyphoid fever is difficult for many health care providers. Although the disease can sometimes be detected through a simple blood culture, a more accurate examination for the disease can be found by a bone marrow culture. Paratyphoid fever and typhoid fever are clinically indistinguishable, thus treatment of the paratyphoid fever is similar to the treatment of typhoid fever. Specific antibiotics can shorten the course of the fever and reduce the chances of death. Paratyphi A infections lead to complications in 10-15% of cases, such as meningitis, endocarditis, hepatic absess, gall bladder cancer, and pancytopenia [2]. Upon infection by Salmonella Paratyphi, the immune system mounts a humoral response predominately producing and utilizing IgA antibodies. Research indicates that a cell-mediated response is also employed. There are currently no vaccines to prevent Salmonella Paratyphi ; however, studies are being conducted concerning the efficacy of the Salmonella Typhi Ty21a-vaccine providing protection from Salmonella Paratyphi as well.
Pathogenesis
The pathogenesis mechanisms of Salmonella Paratyphi bacterium are not completely understood, but there are several key features of the bacterium's ability to infect and colonize a human host. The bacterium is able to survive the low pH of the stomach and enter the intestines. The mechanism which Salmonella Paratyphi uses to adhere to the epithelial cells of the intestines are unknown. Paratyphi B specifically utilizes mechanisms which enable it to cross the intestinal walls and survive in host tissue. A high affinity for iron-uptake utilizing siderophores may also be important regarding the pathogenicity of this particular bacterium [3].
Salmonella Paratyphi initially colonizes the human gastrointestinal tract, causing enteritis, which may or may not result in diarrhea. The bacteria then pass through the mucosa layer and invade the lymphoid tissues and macrophages. If left untreated, Salmonella Paratyphi begin to divide within these cells and are taken to the lymph nodes. A systemic infection develops 8-14 days after infection and the bacteria can now be found in the liver, spleen and bone marrow [19].
Salmonella enterica serovars Paratyphi is transmitted primarily through humans, although there are rare cases of transmission from domesticated animals. The pathogen is most frequently encountered through the ingestion of water, contaminated with feces from an infected individual or an asymptomatic carrier of the disease. Milk, raw vegetables, salads, shellfish, and ice can also transmit the pathogen if not properly washed or prepared. Also, there are rare reports of the disease being transmitted sexually [4].
The infectious dose of Salmonella Paratyphi is generally greater than 1000 organisms. The incubation period of gastroenteritis caused by Salmonella Paratyphi is between one and ten days, however, the incubation period for enteric fever caused by Salmonella Paratyphi is longer, lasting between one and three weeks. Salmonella Paratyphi colonize the intestines, but this can be buffered by stomach acidity. This defense necessitates a greater number of bacteria to be ingested in order to result in symptomatic infection [5].
Paratyphoid fever generally occurs sporadically or in contained outbreaks. Approximately six million cases of Paratyphoid fever are reported annually throughout the world. In the United States approximately one hundred cases are reported, with most occurring in recent travelers. The risk of Paratyphi A is most preeminent in South and Southeast Asia. Southern Asia presents the highest risk of contracting nalixidic acid resistant paratyphoid fever, as well as multidrug resistant paratyphoid fever. Antibiotic resistant strains of paratyphoid fever are resistant to ampicillin, chloramphenicol, and trimethoprim-sulfamethoxazole [4] [5]. Since the start of the millennium, cases of Salmonella enterica serovar Paratyphi, also referred to as Salmonella Paratyphi, have steadly increased [16] [18]. In many areas S. Paratyphi A is now the leading cause of entric fever.
The virulence factors of Salmonella Paratyphi are predominantly the same as those of other Salmonella enterica serotypes. Two of the major virulence factors include the Salmonella pathogenicity Islands I and II. SPI1 encodes genes which contain a type III secretion system. This system allows the pathogen to inject effector proteins directly into the host cell's cytoplasm, and aids in the bacterial invasion of epithelial cells. SPI2 has roles in bacterial replication while within a Salmonella-containing vacuole. SPI2 also encodes for a type III secretion system [6] [7].
Another important virulence factor is found particularly in Paratyphi A. Cytolysin A (ClyA) is a cytotoxic protein which forms pores in membranes. It is a virulence factor most commonly attributed to Escherichia coli, but the gene has been found to be conserved in many Salmonella enterica species [8].
Several studies have been done to investigate different plasmids present in Salmonella enterica serotypes. Their role in virulence is not well understood, but studies have shown that Paratyphi C contains a large cryptic plasmid, which is thought to play some role in the virulence of the strain [6].
It has recently been shown that a fully functional flagellum is required for S. Paratyphi to invade host epithelial cells [17].
Clinical features
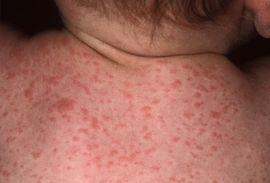
Paratyphoid fever is very similar to typhoid fever. Both fevers are often mistaken for one another. However, symptoms of paratyphoid fever have been known to be less severe because the infection is not nearly as extreme as typhoid fever [18]. Symptoms from paratyphoid fever usually occur between 6-30 days after being infected. The most common, and perhaps prominent, symptom is severe fatigue accompanied by a fever. The fever typically presents as low-grade and can rise as high as 40°C by the third and fourth days of the illness. Additional symptoms that commonly appear include a headache, a distinct red rash over the trunk, cough, stomach pain, loss of appetite, and constipation or severe diarrhea [9] [10] [11].
Once treated, approximately 10% of those with paratyphoid fever will continue to secrete S. paratyphi in their stool for up to three months, which could then be passed on to infect others. Around 2-3% of the recovering patients become permanent carriers for the disease [10]. Paratyphoid fever was once a major cause of mortality throughout the world; however, various prevention mechanisms have begun to eradicate the chance for infection . Nevertheless, the disease is still a problem in tropical and underdeveloped countries of the world, typically Africa, Latin America, and Southeast Asia. Although the findings are some what inconclusive, it is believed that approximately 5.4 million cases of paratyphoid fever occur every year [12].
Diagnosis
The diagnosis of paratyphoid fever can be confirmed by a culture of the blood or bone marrow. As the disease progresses, blood bacterial counts decline which make the blood cultures positive in only about 40-60% of cases. The validity of the test can be increased to 80% if multiple sets of blood cultures are taken. The most accurate way to test for paratyphoid fever is by a bone marrow culture because bacterial counts are typically ten times higher in the bone marrow than in a blood culture. An additional test to diagnose the disease is to perform a biopsy on the rose-colored rash on the trunk of the patient [10].
Treatment
Paratyphoid fever is treated with specific antibiotics to shorten the course of paratyphoid fever and to decrease the chance of death. In most parts of the world ciprofloxacin, a fluoroquinolone, is used to combat the disease. In Indian countries, cases of Paratyphoid fever are more prevalent. In these countries, health officials use cephalosporins for treatment due to increased resistance towards fluoroquinolones and nalidixic acid. Azithromycin has become more commonly used due to the increase of multi-drug resistant strains. After treatment, three to five days may be required for the antibiotic to resolve the fever completely. Some patients may feel worse during the several days of waiting for the fever to end, however, the height of the fever decreases each day. If the fever is not resolved within five days, alternative antibiotics should be considered [13].
Prevention
The incidence of enteric fever is most strongly correlated with poor sanitation and lack of clean drinking water. This is because Salmonella Paratyphi is transmitted through consumption of contaminated food or water. In order to decrease the incidence of this disease it is important to avoid the consumption of contaminated food or water, provide education about food and water safety, and promote basic sanitation [14] [12].
There are currently no vaccines clinically used to defend against Salmonella Paratyphi [12]. Recent studies have indicated that this organism displays evidence drug resistance. For example, it was discovered that Salmonella Paratyphi has limited sensitivity of ofloxacin, nalidixic acid, and ciprofloxacin [14].
Because of the evidence for drug resistance, Salmonella Paratyphi poses a potential health risk and there is a growing necessity for the development of an effective vaccine. There have been studies on the potential use of an oral vaccine that would be used to prevent Salmonella Paratyphi infections, as well as avert typhoid fever. The oral live Salmonella Typhi Ty21a-vaccine has been shown to offer some protection against Salmonella Paratyphi B, slightly less protection against Salmonella Paratyphi A, and no protection against Salmonella Paratyphi C [12]. These varying levels of prevention are due to differences in the shared epitopes of each strain. Despite the research that has been done, there are still no vaccines currently being used in a clinical setting.
Host Immune Response
When the body is infected by Salmonella Paratyphi, a humoral immune response is mounted. The response primarily takes place in the intestinal tract, since the gut is the first line of defense against enteric diseases [15]. Based on research performed using the Ty21a-vaccine, after the host was exposed to the oral vaccine there was a predominance of IgA antibodies. As part of the immune response, it is believed that after the B cells arrive in the intestinal lining, they undergo isotype switching from IgM to IgA. This specific immunoglobulin isotype is produced to stimulate the destruction of Salmonella bacteria. Other research indicates that a cell-mediated response is also activated upon infection [12]. In a 2011 study of a paratyphi outbreak in Napal, researchers found that infected patients had elevated levels of several cytokines, including interferon gamma, tumor necrosis factor alpha (TNF-α), IL-6, IL-8, IL-10, and IL-15. Interestingly, IL-12 was not elevated in patients, but is elevated in patients suffering from nontyphoidal Salmonella infections [19].
References
Created by MaKenzi Burke, Madeline Gabe, Regina Swenton, and Jordan Voth, students of Tyrrell Conway at the University of Oklahoma.
Edited by William Mangin and Tyler Potter, students of Tyrrell Conway at the University of Oklahoma.
Abstract
Background. Enteric fever is a major global problem. Emergence of antibacterial resistance threatens to render current treatments ineffective. There is little research or public health effort directed toward Salmonella enterica serovar Paratyphi A, because it is assumed to cause less severe enteric fever than does S. enterica serovar Typhi. There are few data on which to base this assumption, little is known of the serovar's antibacterial susceptibilities, and there is no readily available tolerable vaccination.
Methods. A prospective study was conducted of 609 consecutive cases of enteric fever (confirmed by blood culture) to compare the clinical phenotypes and antibacterial susceptibilities in S. Typhi and S. Paratyphi A infections. Variables independently associated with either infection were identified to develop a diagnostic rule to distinguish the infections. All isolates were tested for susceptibility to antibacterials.
Results. Six hundred nine patients (409 with S. Typhi infection and 200 with S. Paratyphi A infection) presented during the study period. The infections were clinically indistinguishable and had equal severity. Nalidixic acid resistance, which predicts a poor response to fluoroquinolone treatment, was extremely common (75.25% of S. Paratyphi A isolates and 50.5% of S. Typhi isolates; P
In 2002, there were ∼22 million cases of enteric fever due to infection with Salmonella enterica serovar Typhi, as well as 200,000 deaths [1]. S. enterica serovar Paratyphi A, B, or C is estimated to cause 5.5 million cases of enteric fever each year. However, in some regions—notably, south Asia—the proportion of cases of disease due to S. Paratyphi A, B, or C strains is likely much higher. Despite this, disease due to S. Paratyphi A, B, or C is poorly characterized, with few data regarding risk factors, disease severity, outcome, or antibacterial susceptibilities.
The incidence of enteric fever correlates strongly with poor sanitation and limited access to clean drinking water. Nepal is a developing country, where the disease is endemic and remains the most common clinical and blood culture—confirmed diagnosis among patients with febrile illness [2]. The burden of infection is huge—in Patan Hospital (Kathmandu) alone, we see >2500 suspected cases each year, and the proportion of cases due to S. Paratyphi A increased from 21.7% during 1993–1997 to 33.8% during 1998–2003. This is consistent with reports from elsewhere in Asia that detail an increasing incidence of S. Paratyphi A infection, which is now responsible for up to 15% cases of enteric fever in India and 1% of cases in The Philippines [3–7].
S. Paratyphi A is thought to cause milder disease than does S. Typhi, with predominantly gastrointestinal symptoms [8, 9]. Although this is probably true in the case of S. Paratyphi B infection, there are insufficient data to draw this conclusion for S. Paratyphi A, with a lack of adequately sized clinical studies [10]. The increasing incidence of this infection underlines the need to better characterize its clinical phenotype. The death rate for hospitalized patients with S. Typhi infection varies from 2% in Pakistan and Vietnam to 30%–40% in Indonesia and Papua New Guinea [11]. There are few data from which to generate death or complication rates for S. Paratyphi A infection.
The most important factor in preventing death due to enteric fever is the timely introduction of treatment with effective antibacterials. The emergence of antibacterial resistance has been rapid throughout the treatment history of typhoid and was first reported in 1950 after the introduction of chloramphenicol 2 years previously. In the 1980s and 1990s, S. Typhi developed simultaneous resistance to all first-line drugs, notably chloramphenicol, amoxicillin, and trimethoprim-sulfamethoxazole (TMP-SMZ), encoded on a single plasmid [11]. These multidrug-resistant strains are now widespread, and fluoroquinolones have largely replaced other agents as the drugs of choice. Emerging resistance to fluoroquinolones has been a major setback [12]. In Nepal, the extent of quinolone and fluoroquinolone resistance is uncertain, because most laboratories do not have the facilities for susceptibility testing. Over recent years, we have noticed an increased rate of poor response to first-line treatment (ofloxacin) in patients with enteric fever in our hospital. After fluoroquinolones, treatment choices are limited but include expensive alternatives, such as third-generation cephalosporins or azithromycin [13–19]. Few studies have been done on antibacterial susceptibility patterns of S. Paratyphi A, because it is less frequently the cause of enteric fever. There have been sporadic reports on the growing numbers of drug-resistant S. Paratyphi A in India and Europe [5, 20]. Finally, although reasonably effective vaccines exist for S. Typhi, there are no licensed vaccines available for S. Paratyphi A.
We designed a prospective, descriptive study of enteric fever at Patan Hospital to address the issues regarding etiology, clinical phenotype (according to infecting organism), and antibacterial susceptibilities of infecting isolates.
Materials and Methods
Setting. Kathmandu, the largest city in Nepal, has a population of 1.5 million. Patan Hospital is a 318-bed mission/government hospital providing emergency and elective outpatient and inpatient services. There are 300,000 outpatient visits, 35,000 emergency visits, and 15,000 admissions annually. The vast majority of patients with enteric fever are treated as outpatients.
Patients. The study was conducted from January to August 2004. Ethics approval was obtained from Patan Hospital. All patients of any age who were suspected of having enteric fever and who attended the outpatient clinics or emergency department or who were admitted for treatment were referred to the study physician. After obtaining informed consent, the study physician interviewed and examined each patient, recording the data on a standardized form. Other than culture of blood specimens, laboratory investigations and treatment choice were left to the discretion of the attending physician. Information collected included demographic details, symptoms, physical findings, contact with other cases of enteric fever, recent antibacterial use, and source and use of water purification methods in the patient's home. The study physician also reviewed all blood culture requests and results, so that patients who might have had enteric fever but for some reason had not been referred could be identified and included in the study. For analysis, a patient with enteric fever was defined as any patient with a blood culture positive for S. serovar Typhi or serovar Paratyphi A, B, or C.
Microbiological methods. Blood was inoculated into tryptone soya broth at 37°C, with daily examination for growth over 1 week. Bacteria were identified using standard microbiological tests. Specific antisera distinguished S. Typhi and S. Paratyphi A, B, or C (Murex Diagnostics). Antibacterial susceptibilities were determined at the time of isolation using the modified Kirby-Bauer disk diffusion method, and results were interpreted on the basis of the guidelines for Enterobacteriaceae of the Clinical and Laboratory Standards Institute (formerly the National Committee for Clinical Laboratory Standards) [21]. The antibacterials tested were nalidixic acid, ciprofloxacin, ofloxacin, amoxicillin, chloramphenicol, TMP-SMZ, and ceftriaxone (Himedia Laboratory).
Isolates were stored on brain-heart infusion/glycerol broth at -20°C. MICs were determined for 50 strains of both serotypes using the Etest (AB Biodisk) for nalidixic acid (range of antibacterial concentration in testing, 0.16–256 µg/mL), ciprofloxacin (0.002–32 µg/mL), ofloxacin (0.002–32 µg/mL), gatifloxacin (0.002–32 µg/mL), chloramphenicol (0.16–256 µg/mL), and ceftriaxone (0.002–32 µg/mL).
Data analysis. Statistical analysis was performed using SPSS software for Windows, version 10.0 (SPSS). For categorical or dichotomized parameters, proportions between groups were compared using the χ 2 or Fisher's exact test. For continuous parameters, the Mann-Whitney U test was used. Forward stepwise logistic regression was used to determine significant variables independently associated with either infection. All variables in the univariate analysis with a P value 1
Читайте также: